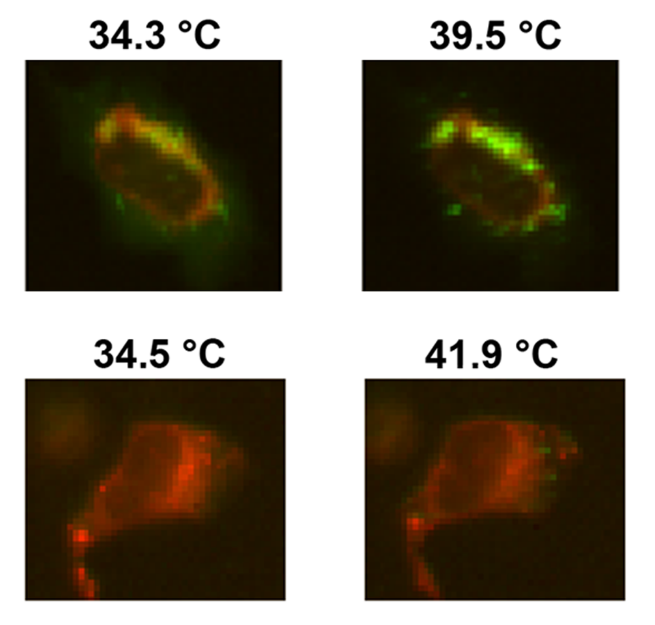
Temperature induced folding of proteins
In the living cell, dynamics of proteins and other biomolecules are affected by the interactions with a myriad of other species present in the cellular matrix. Such interactions are well mapped in vitro but need a more rigorous characterization in-cell to understand the full functionality of the biomolecule.
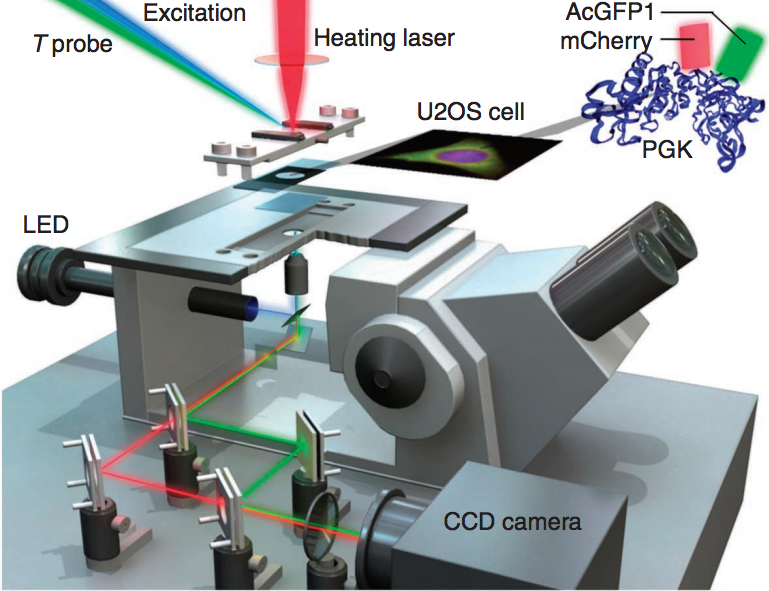
To study folding and unfolding of proteins and other biomolecules in-cells we use Fast Relaxation Imaging (FReI). FReI couples time-resolved fluorescence imaging with fast temperature-jump induced kinetics. Proteins etc. are labeled with a fluorescent probe and are heated using an infrared laser pulse within a few millisecond. The subsequent return to equilibrium after the temperature jump is imaged as a change in FRET between two fluorophores. While labeling a protein with a well known FRET pair, like GFP and mCherry, on the N-terminus and C-terminus allows us to visualize the protein folding in the cell such a system can also be used to study protein-protein and protein-biomolecule interactions by labeling two or more partners with a donor and an acceptor.
S. Ebbinghaus, A. Dhar, J. D. McDonald and M. Gruebele, “Protein folding stability and dynamics imaged in a single cell,” Nature Methods, 7, 319-323 (2010).
Protein-RNA interactions in-cells
Protein-RNA interactions and protein folding are critical subjects in biochemistry, because of their significance during the formation of active complexes and signaling pathways. Regardless of the substantial amount of studies in the fields of protein-RNA interactions and protein folding, little is known about the stability and kinetics of these in the cell. To advance our understanding of protein-RNA interactions inside cells we perform comparative in vivo and in vitro studies, utilizing U1A-SL2 RNA complex and PGK/VlsE proteins as model systems, respectively.
For the protein-RNA studies, dynamics experiments of one positively charged mutant of the spliceosomal U1A protein, the golden model for the RNA Recognition Motif (RRM), revealed a conformational transition for the protein only. Also, U1A-SL2 RNA dissociation kinetics studies with U1A positively charged mutants supported the previously proposed two-step dissociation pathway and demonstrated the importance of positively charge residues. The U1A-SL2 was also investigated in macromolecular crowded buffers were its binding affinity increased. It was also studied inside mammalian cells were it localized in the nucleus and its binding affinity decreased. For the protein folding studies, the extracellular VlsE antigen was found to be destabilized inside mammalian cells as opposed to the intracellular PGK enzyme.
J. Tai, I. Guzman, V. Hahn and M. Gruebele, ” Sub-cellular modulation of protein VlsE stability and folding kinetics,” FEBS. Lett. 590, 1409-1416 (2016).