Phototaxis in E.coli
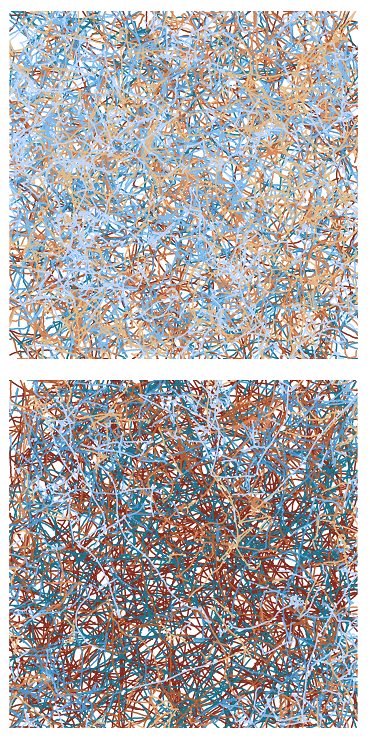
The intricate webs of interwoven lines that you see on the image are formed by overlayed trajectories observed for bacteria E. coli swimming inside the sealed chamber. E. coli swims by alternating two types of motion – ‘runs’, during which cells swim in one direction, and ‘tumbles’, during which cells randomly reorient in one spot. Trajectories shown are colored according to their ‘tumbliness’: brighter colors correspond to trajectories with more tumbles. Remarkably, E. coli is able to migrate to more favourable environment solely by modulating the fraction of tumbles in response to changes in the external conditions essential for the survival of bacteria such as chemical composition, temperature, pH etc. The goal of this research project is to study how light affects E. coli motile behavior. Turns out that E. coli responds to high intensity blue light by decreasing the fraction of tumbles or prolonged running. The effect is especially obvious if you compare trajectories of the illuminated bacteria shown on the top with those on the bottom. This result suggests that E. coli will migrate up the gradient of blue light and accumulate in more illuminated areas, although the adaptive value of this response remains to be found.
We are especially interested in studying phototaxis in E. coli on the population level in heterogeneous illumination environment that we can create using spatial light modulator (SLM). Not only that but the ability to manipulate light gradients at will using SLM gives us an instrumental tool to regulate the distribution and density of phototaxing E. coli. Eventually we plan to use this technique to study chemosensing coordination in E. coli, i.e. how quorum-sensing is coupled to bacterial motility.
Zebrafish locomotion
How simple is the underlying control mechanism for the complex locomotion of vertebrates? We explore this question for the swimming behavior of zebrafish larvae. Current research usually uses arbitrarily selected criteria to distinguish and classify zebrafish behaviors. Our lab has established a parameter-independent method to characterize zebrafish swimming in a more quantitative way.
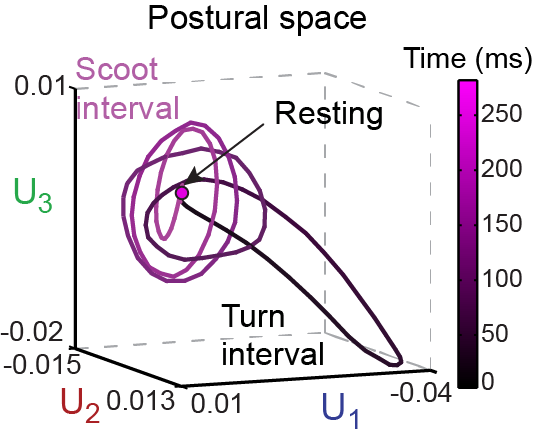
The fish motion itself yields a natural set of fish “eigenshapes” as coordinates, rather than the experimenter imposing a choice of coordinates. Three eigenshape coordinates are sufficient to construct a quantitative “postural space” that captures >95% of the observed zebrafish locomotion. Viewed in postural space, swim bouts are manifested as trajectories consisting of cycles of shapes repeated in succession. To classify behavioral patterns quantitatively and to understand behavioral variations among an ensemble of fish, we construct a “behavioral space” using multidimensional scaling (MDS). This method turns each cycle of a trajectory into a single point in behavioral space, and clusters points based on behavioral similarity.
Furthermore, with the insight into fish behavior from postural space and behavioral space, we construct a two channel neural network model for fish locomotion, which produces comparable postural space and behavioral space dynamics compared to real zebrafish.
K Girdhar, M Gruebele, Y. R. Chemla, “The behavioral space of zebrafish locomotion and its neural network analog”, PLOS ONE, 10(7): e0128668 (2015).