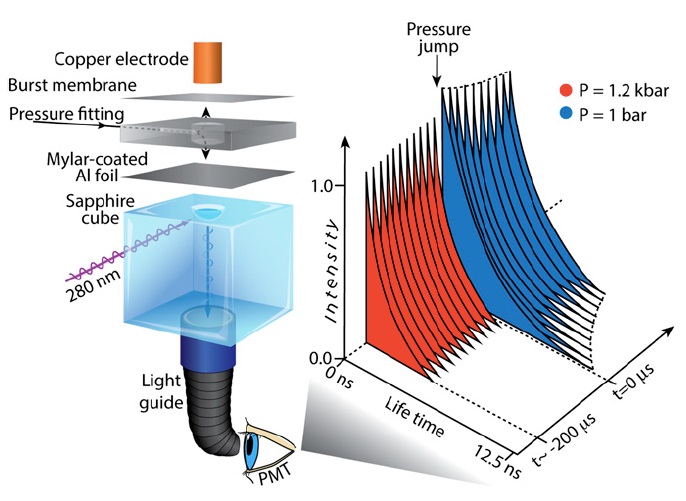
Pressure induced protein folding
Pressure is an alternative to temperature and denaturants to perturb protein structure and probe protein folding. Pressure-jump probed folding kinetics are a useful experiment to compare to molecular dynamics simulations of protein folding dynamics.
The challenge of doing pressure jump experiments on fast-folding proteins is time-resolution. The Gruebele group has developed a laser-probed pressure-jump apparatus that can deliver up to 3000 bar pressure drops with ~1 μs resolution, currently the largest and fastest pressure jumps in the field. We have measured the pressure-probed kinetics of two fast folders, lambda repressor and the WW-domain, and collaborated with Klaus Schulten’s group to model the pressure-induced refolding of the proteins in long, all-atom molecular dynamics simulations.
A. J. Wirth, Y. Liu, M. Prigozhin, K. Schulten and M. Gruebele, ” Comparing Fast Pressure and Temperature Jump Protein Folding by Experiment and Simulation,” JACS 137, 7152-7159 (2015).
Protein-RNA interactions in vitro
In genes, the protein-coding sections of DNA are interrupted by non-coding sections of DNA, which must be removed by a process called precursor mRNA (pre-mRNA) splicing in order for the gene to work. The spliceosome, a macromolecular machine, performs this splicing. Disruption of pre-mRNA splicing is a common feature of numerous genetic diseases including cancer, metabolic, and neurodegenerative diseases. Detailed studies of spliceosome assembly will determine how conformational and compositional changes dictate splicing outcomes, and highlight assembly changes that lead to pathological conditions.
Using a combination of stopped flow, T-jump, equilibrium assays, and fluorescence titrations, we probe the kinetics and binding affinities of the RNA binding protein U1A and its binding partner, stem loop 2 (SL2). Currently, fluorescence titrations utilizing FRET and intrinsic tryptophan fluorescence monitor binding affinity to complement in vivo experiments and to determine the relative role of positively charged residues in U1A.
D. Anunciado, A. Dhar, M. Gruebele, and A. M. Baranger, “Multistep Kinetics of the U1A-SL2 RNA Complex Dissociation,” Journal of Molecular Biology, 5, 896–908 (2011).